These are the research projects currently ongoing in the lab. If you think you have similar interests and would like to collaborate, please reach out!
Understanding the demographic history of ancient domesticated populations
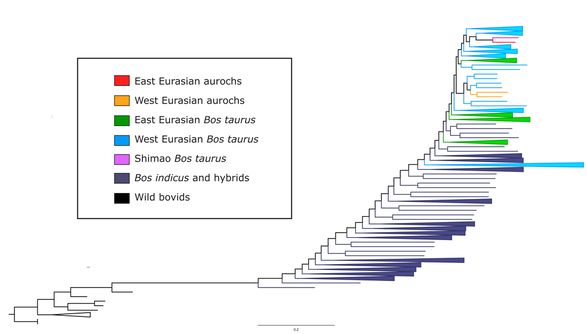
As ancient human populations moved throughout the globe, they took their domesticated animals with them. By studying ancient animal populations, we learn more about how these animals were used, and can use them as a proxy to understand human migrations and interactions. Current and ongoing projects are focused on two areas of research: the population history of domestic dogs in the Americas before European colonization, and interactions between domesticated cattle and wild aurochs in China and Mongolia. We are interested in reconstructing the history of migration, admixture, and selection in these populations, and understanding what this means for the humans that managed these animals.
As ancient human populations moved throughout the globe, they took their domesticated animals with them. By studying ancient animal populations, we learn more about how these animals were used, and can use them as a proxy to understand human migrations and interactions. Current and ongoing projects are focused on two areas of research: the population history of domestic dogs in the Americas before European colonization, and interactions between domesticated cattle and wild aurochs in China and Mongolia. We are interested in reconstructing the history of migration, admixture, and selection in these populations, and understanding what this means for the humans that managed these animals.
Clarifying archaic gene flow events using modern human genomes
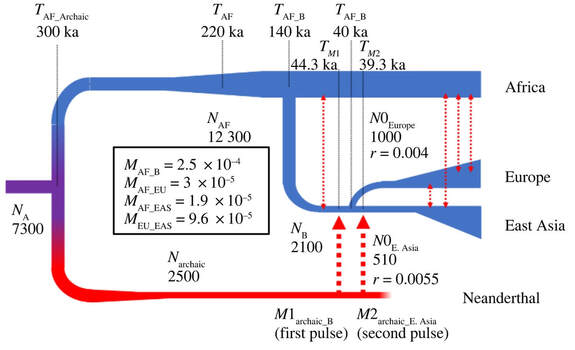
We know that most modern human populations have ancestry from Neanderthals and Denisovans, but there is ongoing debate about where and how often modern humans encountered archaic humans. We can use the archaic ancestry still present in living humans to learn more about the number, place, and timing of archaic introgression events with the help of demographic simulations. These simulations are useful for not only understanding past modern and archaic human interactions, but also for learning more about population structure and the demographic history of Neanderthals and Denisovans.
We know that most modern human populations have ancestry from Neanderthals and Denisovans, but there is ongoing debate about where and how often modern humans encountered archaic humans. We can use the archaic ancestry still present in living humans to learn more about the number, place, and timing of archaic introgression events with the help of demographic simulations. These simulations are useful for not only understanding past modern and archaic human interactions, but also for learning more about population structure and the demographic history of Neanderthals and Denisovans.
Examining impacts of archaic introgression on modern human health
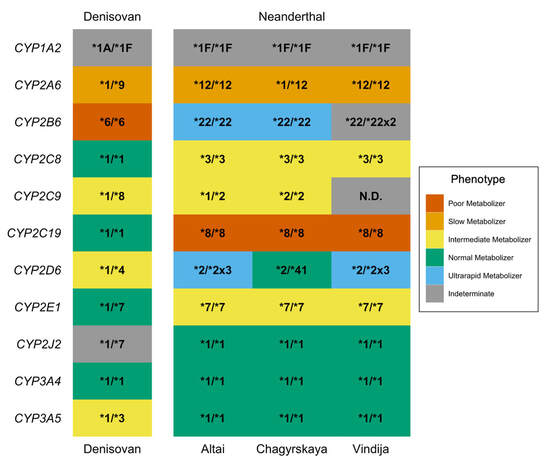
As modern humans encountered new environments, some of the variants they inherited from archaic humans were selected for because they were adaptive. However, not all archaic variants have a positive impact on human health and fitness - others can be maladaptive and cause disease in living individuals. We are interested in exploring genetic variants that are archaic in origin and show signals of selection in modern populations, and then characterizing how those archaic variants affect gene function and health. We are also interested in genetic variants that are at higher frequency in specific geographic regions or populations, as they may represent "local adaptations" - genetic variants that helped a population adapt to a specific environment.
As modern humans encountered new environments, some of the variants they inherited from archaic humans were selected for because they were adaptive. However, not all archaic variants have a positive impact on human health and fitness - others can be maladaptive and cause disease in living individuals. We are interested in exploring genetic variants that are archaic in origin and show signals of selection in modern populations, and then characterizing how those archaic variants affect gene function and health. We are also interested in genetic variants that are at higher frequency in specific geographic regions or populations, as they may represent "local adaptations" - genetic variants that helped a population adapt to a specific environment.